 This post was first published at ScraperWiki.
This post was first published at ScraperWiki.
Regular readers of this blog will know I’ve read quite few machine learning books, now to put this learning into action. We’ve done some machine learning for clients but I thought it would be good to do something I could share. The Forest Cover Type Prediction challenge on Kaggle seemed to fit the bill. Kaggle is the self-styled home of data science, they host a variety of machine learning oriented competitions ranging from introductory, knowledge building (such as this one) to commercial ones with cash prizes for the winners.
In the Forest Cover Type Prediction challenge we are asked to predict the type of tree found on 30x30m squares of the Roosevelt National Forest in northern Colorado. The features we are given include the altitude at which the land is found, its aspect (direction it faces), various distances to features like roads, rivers and fire ignition points, soil types and so forth. We are provided with a training set of around 15,000 entries where the tree types are given (Aspen, Cottonwood, Douglas Fir and so forth) for each 30x30m square, and a test set for which we are to predict the tree type given the “features”. This test set runs to around 500,000 entries. This is a straightforward supervised machine learning “classification” problem.
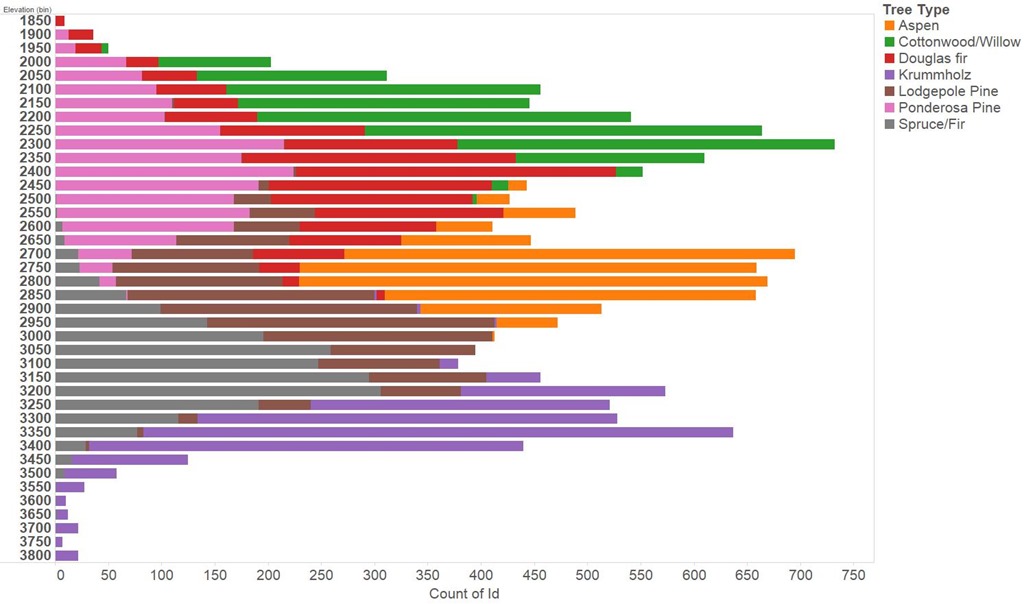
The first step must be to poke about at the data, I did a lot of this in Tableau. The feature most obviously providing predictive power is the elevation, or altitude of the area of interest. This is shown in the figure below for the training set, we see Ponderosa Pine and Cottonwood predominating at lower altitudes transitioning to Aspen, Spruce/Fir and finally Krummholz at the highest altitudes. Reading in wikipedia we discover that Krummholz is not actually a species of tree, rather something that happens to trees of several species in the cold, windswept conditions found at high altitude.
Data inspection over I used the scikit-learn library in Python to predict tree type from features. scikit-learn makes it ridiculously easy to jump between classifier types, the interface for each classifier is the same so once you have one running swapping in another classifier is a matter of a couple of lines of code. I tried out a couple of variants of Support Vector Machines, decision trees, k-nearest neighbour, AdaBoost and the extremely randomised trees ensemble classifier (ExtraTrees). This last was best at classifying the training set.
The challenge is in mangling the data into the right shape and selecting the features to use, this is the sort of pragmatic knowledge learnt by experience rather than book-learning. As a long time data analyst I took the opportunity to try something: essentially my analysis programs would only run when the code had been committed to git source control and the SHA of the commit, its unique identifier, was stored with the analysis. This means that I can return to any analysis output and recreate it from scratch. Perhaps unexceptional for those with a strong software development background but a small novelty for a scientist.
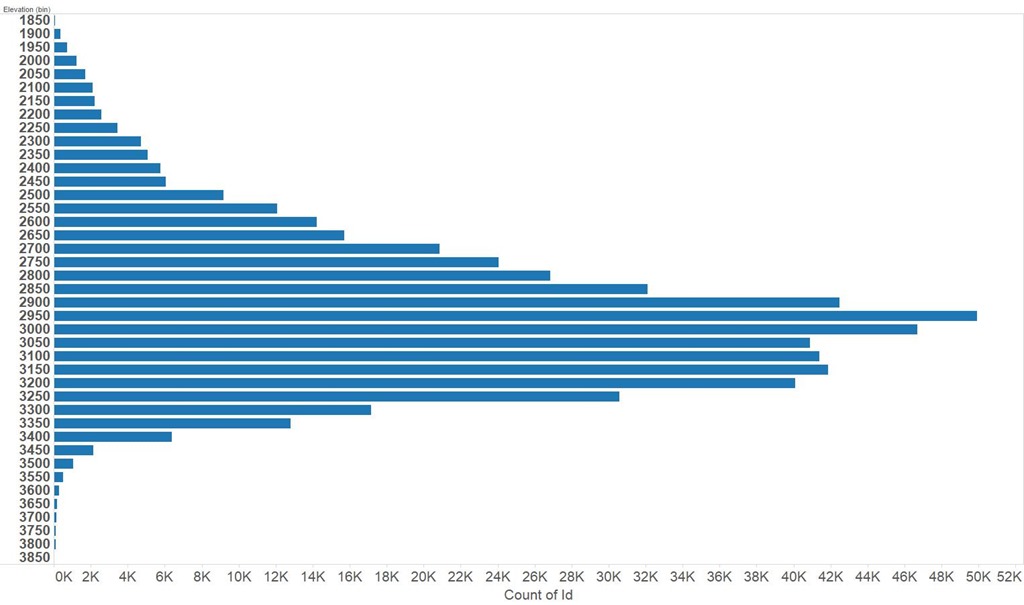
Using a portion of the training set to do an evaluation it looked like I was going to do really well on the Kaggle leaderboard but on first uploading my competition solution things looked terrible! It turns out this was a common experience and is a result of the relative composition of the training and test sets. Put crudely the test set is biased to higher altitudes than the training set so using a classifier which has been trained on the unmodified training set leads to poorer results then expected based on measurements on a held back part of the training set. You can see the distribution of elevation in the test set below, and compare it with the training set above.
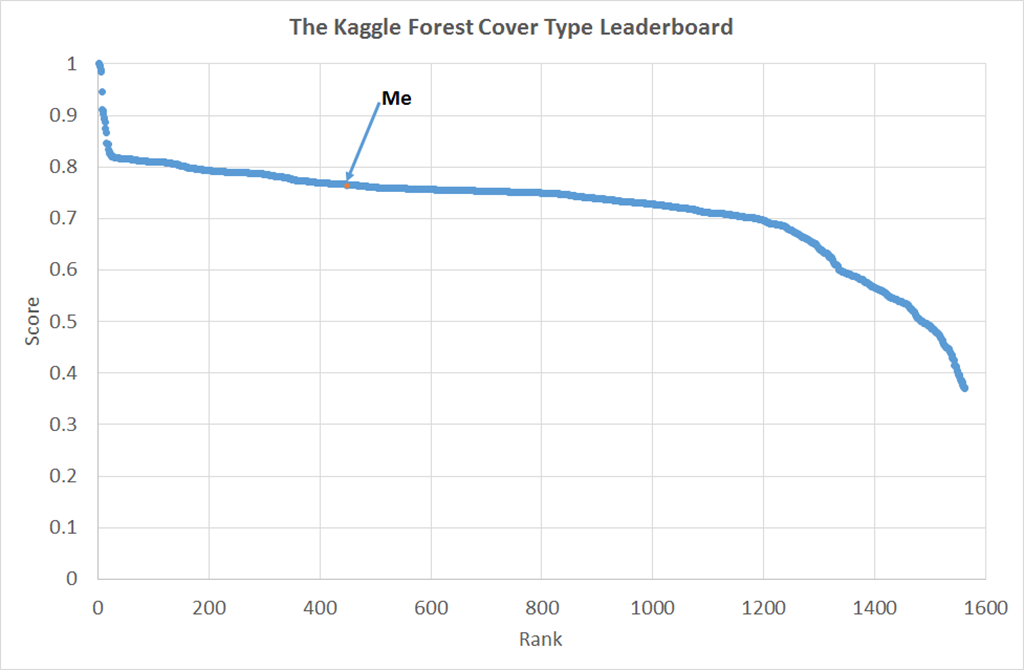
We can fix this problem by biasing the training set to more closely resemble the test set, I did this on the basis of the elevation. This eventually got me to 430 rank on the leaderboard, shown in the figure below. We can see here that I’m somewhere up the long shallow plateau of performance. There is a breakaway group of about 30 participants doing much better and at the bottom there are people who perhaps made large errors in analysis but got rescued by the robustness of machine learning algorithms (I speak from experience here!).
There is no doubt some mileage in tuning the parameters of the different classifiers and no doubt winning entries use more sophisticated approaches. scikit-learn does pretty well out of the box, and tuning it provides marginal improvement. We observed this in our earlier machine learning work too.
I have mixed feelings about the Kaggle competitions. The data is nicely laid out, the problems are interesting and it’s always fun to compete. They are a great way to dip your toes in semi-practical machine learning applications. The size of the awards mean it doesn’t make much sense to take part on a commercial basis.
However, the data are presented such as to exclude the use of domain knowledge, they are set up very much as machine learning challenges – look down the competitions and see how many of them feature obfuscated data likely for reasons of commercial confidence or to make a problem more “machine learning” and less subjectable to domain knowledge. To a physicist this is just a bit offensive.
If you are interested in a slightly untidy blow by blow account of my coding then it is available here in a Bitbucket Repo.




 I’ve discovered that my blog is actually a good place to put things I need to remember see, for example, my blog post on running
I’ve discovered that my blog is actually a good place to put things I need to remember see, for example, my blog post on running